Induced pluripotent stem cells (iPSCs) have recently emerged as a promising avenue for advancing allogeneic cell therapies. Through the integration of gene-editing technologies, iPSCs offer distinct advantages over conventional donor-derived allogeneic cell therapies. Yet the process of introducing multiple genetic alterations using CRISPR technology presents challenges, particularly within sensitive iPSC environments. Efficient delivery of CRISPR components proves challenging in iPSCs due to their lower transfection and editing efficiencies compared to other cell types. Furthermore, it is imperative to maintain the pluripotent state of iPSCs throughout the gene-editing process. In addition, isolating individual cells and establishing clonal cell populations with desired genetic modifications presents a significant hurdle in genome-editing procedures for iPSCs. This article summarizes the challenges of employing gene-editing techniques for the development of iPSC-derived allogeneic cell therapies.
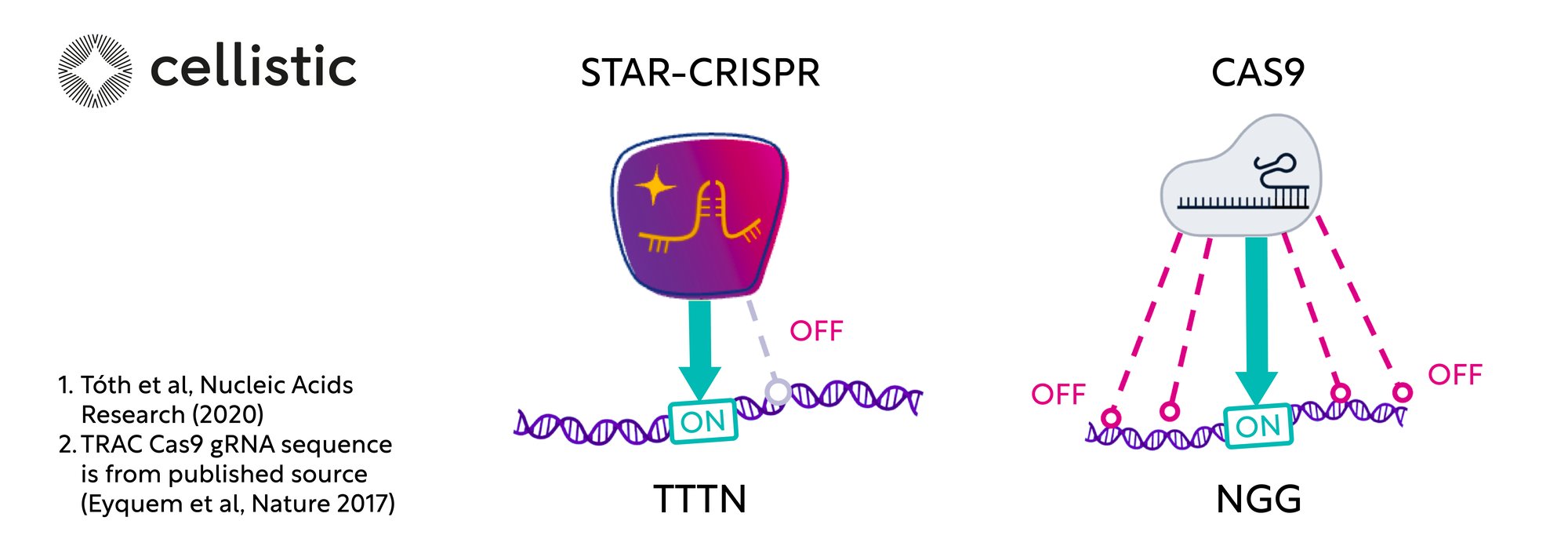
One of the primary decisions in gene editing is the choice of a CRISPR system between two commonly used classes: type II and type Va nuclease-based systems. Cas9 and Cas12a are the most commonly used nucleases representing those systems, each offering distinct advantages. Cas9, with its well-established track record, provides versatility and efficiency in inducing targeted double-strand breaks (DSBs). On the other hand, Cas12a exhibits enhanced specificity and flexibility due to its unique PAM recognition sequence and lower off-target effects. In recent years, engineered variants of Cas9 and Cas12 – including both high-fidelity (HiFi) or enhanced specificity (eSp) Cas9 and STAR nucleases – have been developed. These variants feature amino acid modifications that enhance their DNA cleavage efficiency, versatility, and precision.
Delivery methods introduce a key variable in the success of gene editing, as well. While traditional approaches using plasmids or lentiviral vectors (LVVs) offer convenience, newer techniques involving ribonucleoprotein (RNP) complexes provide several advantages, including transient expression, reduced immunogenicity, and enhanced precision. This transient nature decreases the window of opportunity for off-target effects to occur, as the CRISPR components are present in the cell for less time.
Selecting the right target sequence for guide RNA design is another crucial aspect influencing the specificity of gene editing. Ensuring perfect homology with the target site minimizes off-target effects, safeguarding against unintended genomic alterations. Furthermore, it's important to consider avoiding targeting exons with multiples of three nucleotides for knockouts and avoiding the last exon before the stop codon. This strategy allows for nonsense-mediated decay (NMD) to occur, effectively disrupting the open reading frame (ORF) of the targeted gene and preventing protein production.
For knock-in strategies utilizing homology-directed repair (HDR), careful selection of the type of HDR template and optimizing the distance from the cut site are paramount: A well-designed template with precise homology arms and appropriate length ensures efficient and site-specific integration of exogenous DNA sequences into the genome.
The importance of single-cell cloning in gene editing cannot be overstated. By isolating and characterizing individual edited cells, clones with desired genomic modifications can be identified and selected, ensuring the reproducibility and consistency of therapeutic outcomes. Additionally, it ensures that the clones derived from a single cell possess the intended gene edits while avoiding any undesired (off-target) modifications.
Typically, generating iPSC-derived off-the-shelf allogeneic cell therapies requires approximately 4-6 gene edits on average. Each round of gene editing involves culturing cells for a minimum of three months and when performing four gene edits sequentially, cells must therefore be cultured for at least one year. However, it's important to note that iPSCs, like any other cell, are susceptible to culture-acquired genetic alterations. To address this challenge, implementing multiplex editing becomes crucial, as it involves optimizing gene-editing protocols in such a way to minimize the number of clones that require screening to identify the iPSC clone(s) of interest, and, in turn, reduce manufacturing costs.
In conclusion, the success of iPSC allogeneic cell therapy hinges on the meticulous implementation of advanced gene-editing strategies. By harnessing the power of CRISPR nucleases, employing single-cell cloning techniques, selecting optimal delivery methods, and refining guide RNA design, precision, safety, and efficacy in modifying iPSCs for therapeutic applications can be enhanced.
Author: Suzanne Snellenberg

